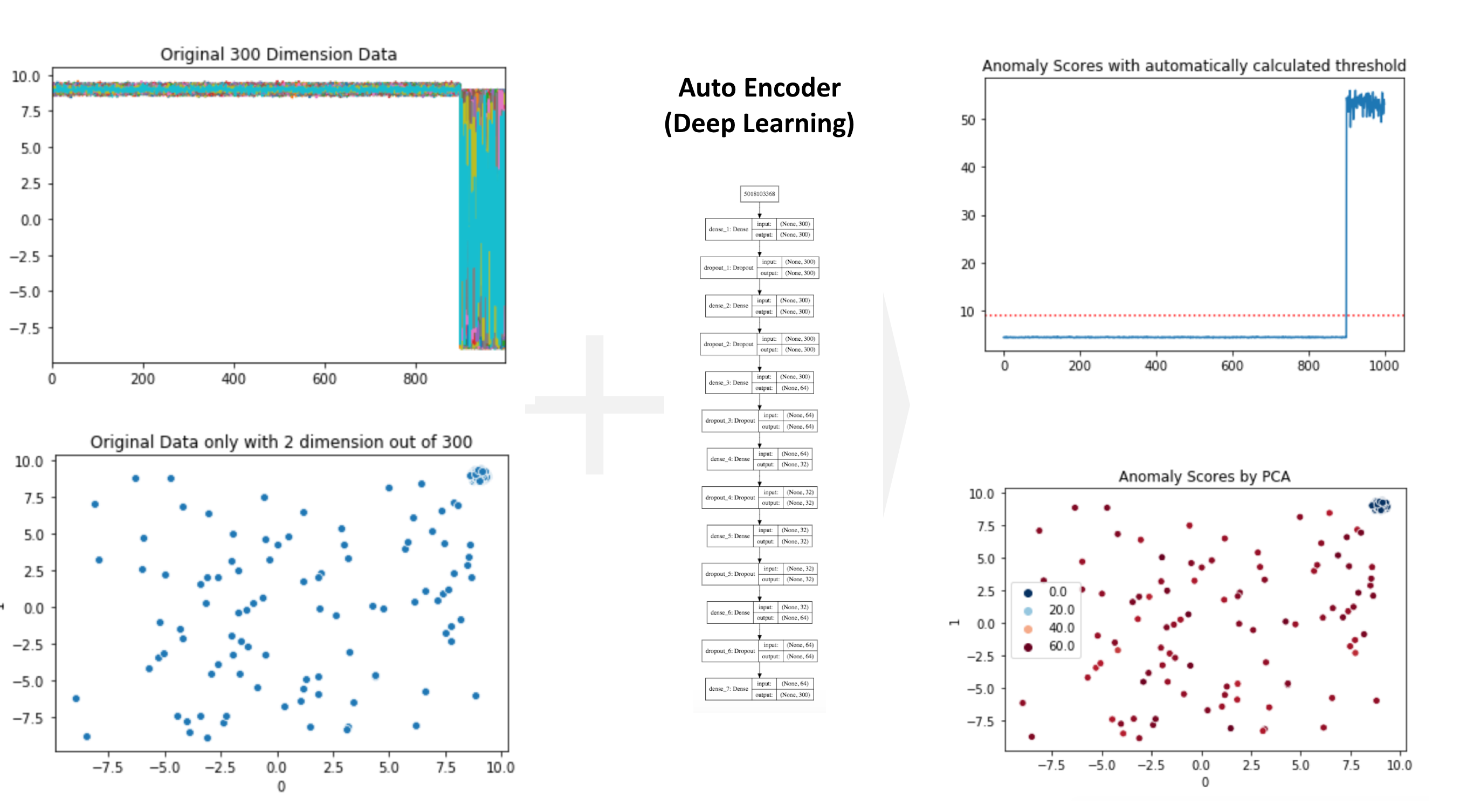
Loading Audio Data
Libraries¶
import pandas as pd
import numpy as np
from scipy.io import wavfile
import matplotlib.pyplot as plt
%matplotlib inline
Load a file as numpy¶
filename = '../data/sound00.wav'
fs, data = wavfile.read(filename)
data
Visualize audio data¶
plt.figure(figsize=(16, 4))
plt.plot(data, lw=0.5, alpha=0.8);